(Epi)Genetic Profiling Services
Overview
Please Note:
We have changed our sample drop off protocols and times - click on and read the following PDFs for details:
- The core accepts samples that require library preparation and sequencing. There will be a quality control step prior to agreement of service. Please refer to specific services using the menu on the left for detailed quality control descriptions.
- The price for library preparation is per library, and includes sample quality control, library preparation and library validation.
- The price for sequencing varies based on sample volume and number of reads required per sample, and includes de-multiplexing (if required), post-processing (if available and requested) and two years data storage. For methylation sequencing services, methylation calls are also provided with alignments as part of the post-processing.
- Sequencing is performed on NovaSeq 6000 or MiSeq Illumina instruments.
- A Bioinformatics Fee (10% of the sequencing price for Internal Clients and 20% for External Clients) will be added.
- Libraries made by the core, routinely yield clusters between 750-800 k/mm2 passing the illumina chastity filter. We cannot guarantee similar clustering and/or quality for libraries made by customers of the core.
- We require that core clients acknowledge the Epigenomics Core of Weill Cornell Medical College in publications and presentions enabled by Epigenomics core resources.
Sample submission tips
- To request a sequencing service, please fill out a request in our Agilent Crosslab/iLab Service Request LIMS. Please refer to the GETTING STARTED section of our Sample Workflow above for detailed sample submission instructions.
- Please use the Agilent Crosslab/iLab service ID and the sample number you have indicated in the submission form to label your tubes clearly, tubes without a service ID and sample number will not be accepted.
- Once samples are submitted and pass quality control, they are entered into a sample or library queue.
- The MiSeq personal sequencer has a single lane and only supports paired end clustering. [MiSeq PDF]
Timeline:
Approximate time for library preps and sequencing is 4-8 weeks if all samples pass quality control.
Approximate time for data pre-processing and transfer is 1-2 weeks from the end of a successful sequencing run.
RRBS (DNA Methylation Sequencing)
Assay Description
Reduced Representation Bisulfite Sequencing is a modification of the original RRBS protocol (Gu H. et al. 2011) and the in-house developed ERRBS method (Akalin A., Garrett-Bakelman F. et al, 2012) for base-pair resolution methylation sequencing analysis based on the use of a restriction enzyme to enrich for CpG fragments. RRBS starts with MspI digestion, followed by NGS-library preparation and bisulfite conversion of cytosines.
Our RRBS protocol yields about ~10% of genomic CpG sites (roughly 3M CpGs in the human genome), and provides enrichment in CpG islands and CpG shores, promoters, exons, introns and intergenic regions (Garrett-Bakelman F., Sheridan, C. et al. 2015).
This assay requires RNA-free high molecular weight DNA. Degraded DNA, such as that obtained from FFPE is not suitable. FFPE samples may be processed using Agilent's Methylome Capture assay instead.
Sample Requirement:
Submit 75 ng of genomic DNA, of molecular weight >20kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Agilent Tape station 4200 or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
100 million (M) read per sample on a paired end read flow cell with 100bp read length (PE100) are recommended for differential methylation analysis, for detailed information please review Garrett-Bakelman F., Sheridan, C. et al. 2015.
Third Party Resource: RRBS Guide from Babraham Bioinformatics (makers of the Bismark aligner, FASTQC, Seqmonk etc.)
Some studies that have used our RRBS assay: Odell SC. et al. 2020, Emi T. et al. 2020
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep | $200 | $260 |
| NextSeq 2000 (400M reads) Single Read 100bp (SR100) + Bioinformatics Processing | $1980 | |
| NovaSeq X (1500M reads) Single Read 100bp (SR100) + Bioinformatics Processing | $3484 |
Prices may vary depending on the number of samples.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
RR(Ox)BS (5hmC Sequencing)
Assay Description
Reduced Representation Oxidative Bisulfite Sequencing enables an accurate 5-hydroxymethylcytosine (5hmC) identification. Please note that the protocol requires two library preparations per sample. The core uses methodology described in Akalin A., Garrett-Bakelman F. et al, 2012 and Garrett-Bakelman F., Sheridan, C. et al. 2015), with the chemistry from Tecan’s Ultralow Methyl-Seq with TrueMethyl oxBS to prepare the libraries.
Sample Requirement:
Submit 400 ng of genomic DNA, of molecular weight >20kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul. Due to sequencing recipe requirements please submit a minimum of 4 samples (8 libraries).
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Agilent Tape station 4200 or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
100 million (M) reads per library on a single end read flow cell with 100 sequencing cycles (SR100) are recommended for differential methylation analysis, for detailed information please review Garrett-Bakelman F., Sheridan, C. et al. 2015.
Some studies that have used our RRoxBS assay: Singh P. et al. 2021, Fortin J. et al. 2023
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep (per sample) | $250 | $350 |
| NextSeq 2000 (400M reads) Single Read 100bp (SR100) + Bioinformatics Processing | $1980 | |
| NovaSeq X (1500M reads) Single Read 100bp (SR100) + Bioinformatics Processing | $3484 |
Prices may vary depending on the number of samples.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
Whole Genome Bisulfite Sequencing (WGBS) - Genome-wide methylation sequencing
Assay Description
Whole genome bisulfite sequencing is the gold-standard approach for comprehensive base-pair resolution and quantitative information at most genomic cytosines. The core uses the Accel-NGS Methyl-Seq DNA library kit to prepare libraries. This method uses cytosine bisulfite conversion followed by single stranded ligation of adapters and PCR amplification. [Application Note].
Sample Requirement:
Submit 300 ng of genomic DNA, of molecular weight >20kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Agilent Tape station 4200 or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep | 250 | 325 |
| NovaSeq X (1500M reads) Paired End 150bp (PE150) + Bioinformatics Processing | 4500 | |
| NovaSeq X (10000M reads) Paired End 150bp (PE150) + Bioinformatics Processing | 16800 |
To calculate the sequencing depth required for your desired coverage and species of interest, please use the Illumina coverage calculator.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
Whole Genome Oxidative Bisulfite Sequencing (WGoxBS) - Genome-wide 5hmC sequencing
Assay Description
Whole Genome Oxidative Bisulfite Sequencing (WGoxBS) enables an accurate 5-methylcytosine (5mC) identification and interrogation of both 5-hydroxymethylcytosine (5hmC) and 5-methylcytosine (5mC). Please note that the protocol requires two library preparations per sample, and therefore twice the sequencing depth than WGBS. The core uses Tecan’s Ultralow Methyl-Seq with TrueMethyl oxBS kit to prepare libraries.
Sample Requirement:
Submit 400 ng of genomic DNA, of molecular weight >20kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul. Due to sequencing recipe requirements please submit a minimum of 6 samples (12 libraries).
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Agilent Tape station 4200 or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep | 320 | 400 |
| NovaSeq S2 (6 samples) pair end 150 cycles (2x150) + Bioinformatics Processing | 14960 | 16000 |
To calculate the sequencing depth required for your desired coverage and species of interest, please use the Illumina coverage calculator.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
Methylome Capture Sequencing (targeted methylome sequencing)
Assay Description
Methylcapture is a hybridization-based approach on platforms containing pre-designed capture oligos, followed by methylation sequencing. There are commercially designed capture libraries available with a range
of epigenetic features that cover ~12% to ~24% of all human genome CpGs [Agilent SureSelect MethylSeq | Roche CpGiant].
Please contact us if you would like to use this service.
Advantages:
- Researchers can custom-design capture libraries that can be used for validation or for discovery of novel epigenomic regions. Please contact us if you would like to design a custom library.
- Amenable for FFPE samples. Since the technique relies on a sonication step, FFPE DNA can be used.
Sample Requirement:
Depending on the platform used, submit 0.5-2ug of genomic DNA, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2, RNA-free and at a concentration of ~50ng/ul
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Perkin Elmer Labchip GX or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep | 400 | 500 |
PE100 (2x100 cycles) sequencing is recommended. Depth depends on the chosen commercially available capture panel. To calculate the sequencing depth required for you panel/species, please use the Illumina coverage calculator.
The same data processing pipeline we use for our RRBS assay will be used for methylome capture data as well.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
Enzymatic Methyl-Seq (EMSeq) - Genome-wide methylation sequencing
Assay Description
The Enzymatic Methyl Sequencing assay provides a high-performance enzyme-based alternative to bisulfite conversion for identification of methylation in low input samples. The core uses the NEBNext Enzymatic Methyl-seq kit to prepare libraries.
Sample Requirement:
Submit 10-50 ng of genomic DNA, of molecular weight >20kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul.
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Agilent Tape station 4200 or agarose gel to determine molecular weight.
[Click here for a detailed description of the QC]
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Library Prep | 220 | 300 |
| NovaSeq X (1500M reads) Paired End 150bp (PE150) + Bioinformatics Processing | 4500 | |
| NovaSeq X (10000M reads) Paired End 150bp (PE150) + Bioinformatics Processing | 16800 |
To calculate the sequencing depth required for your desired coverage and species of interest, please use the Illumina coverage calculator.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
We process Bisulfite sequencing data using an in-house BisSeq pipeline (Garrett-Bakelman F., Sheridan, C. et al. 2015) that generates 12 files for each sample:
methylcall.CpG.Sample.1x.txt.gz
methylcall.CHG.Sample.1x.txt.gz,
methylcall.CHH.Sample.1x.txt.gz,
cgunits.Sample.1x.txt.gz,
The *.1x.txt.gz files contain all reported sites for CpG, CHG, and CHH contexts. The minimum read coverage cutoff for this file is 1
The cgunits file contains the reported sites for CpG context where consecutive CpGs for the forward and reverse strand have been combined into one site, or CpG-unit.
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are as follows:
- chrBase = This is the name (chromosome.base location)
- chr = chromosome on which the methylated base is located
- base = location of methylated base on the chromosome
- strand = forward strand (F) or reverse strand (R)
- coverage = read coverage
- freqC = % methylated
- freqT = % unmethylated
cpg.Sample.10x.txt.gz,
chg.Sample.10x.txt.gz,
chh.Sample.10x.txt.gz
The *.10x.txt.gz file contain sites with greater than or equal to 10x coverage for CpG, CHG, and CHH contexts
The minimum read coverage cutoff for this file is 10
These are tab delimited text files that contain the locations of C's in either CpG, CHG, or CHH, context and their methylation levels
The column headers are the same as above.
Sample_CpG.wig - This is a wiggle file that allows the display of the methylation levels of CpG sites at their location in the genome in a track format for uploading into a genome browser such as the UCSC genome browser or Broad Institute's IGV.
Sample.bam - This file contains the complete alignments in binary (BAM) format as output by the Bismark bisulfite mapper.
Sample.bedGraph.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
It is a sorted bedGraph file that reports the position of a given cytosine and its methylation state.
The bedGraph output is tab delimited with 0-based start coords and 1-based end coords. The columns are:
track type=bedGraph
chromosome
start position
end position
methylation percentage
Sample.bismark.cov.gz - This file contains methylation information for individual cytosines as output by the Bismark methylation extractor.
The coverage output is tab delimited with 1-based genomic coords and the columns are:
chromosome
start position
end position
methylation percentage
count methylated
count non-methylated
Sample_summary.txt - This file summarizes adapter trimming, alignment information, and mapping efficiency of the sample against the genome, in addition to the conversion rate and methylation statistics based on all reported CpG sites (cutoff of 10x), such as the average and median conversion rates plus the number of CpG's covered.
5hmC-BIC-Seq (detection of 5-hydroxymethylcytosine modification by affinity pull down)
Assay Description
5hmC-bead-integrated-click-sequencing is an in-house method developed by the core to profile 5hmC containing DNA sequences on a genome-wide scale that uses a novel integrated approach. Typical enrichment protocols use antibodies that recognize a modification or set of modifications on DNA. We chose a covalent chemical labeling technique that could be integrated into the NGS library preparation process. DNA sequences with 5hmC moieties are directly modified with azide-glucose, which can then form a stable biotin conjugate through bio orthogonal click-chemistry. Streptavidin affinity purification enriches 5hmC-containing DNA sequences and integration with sample preparation steps creates a robust assay that can accept limiting levels of input DNA.
Please contact us if you wish to use this service.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
ChIPseq (genome-wide mapping of DNA binding proteins)
Assay description
Chromatin Immunoprecipitation Sequencing (Carey et al, 2009 http://cshprotocols.cshlp.org/) is the primary method for profiling protein DNA interactions, based on the enrichment of DNA associated to a protein of interest or a histone modification. Despite its increasing use ChIP-seq libraries are among the most challenging to perform and require significant experience and quality control measures. The core offers library preparation for chromatin immunoprecipitated material after successful quality control.
Sample Requirement:
For Input material submit 50 ng at a concentration of 5ng/ul.
For ChIP material submit ~22ng of DNA at a concentration of 0.3ng/ul to 1ng/ul.
Please use a double stranded fluorometric method to determine concentration (Qubit for example); concentrations determined by nanodrop are not reliable for this assay.
Epigenomics Core Quality Control:
QC1 - Quantity: dsDNA using Qubit Fluorometer.
QC2 - Quality: High Sensitivity DNA Bioanalyzer chip to determine the spread of the Input chromatin. Accurate representation of the original ChIPd material is obtained when the size range of the DNA fraction required for library preparation (130-230bp) is > 10% of the total DNA provided.
[Click here for a detailed description of the QC]
Library preparation for this assay is currently done using the IDT-xGen kit.
Some studies that have used our ChIPSeq sequencing service: Popovic R. et al. 2014, Kuo PY. et al. 2014, Qiao Y. et al. 2013
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| IDT-xGen ChIP-seq Library Prep | 175 | 225 |
| NextSeq 2000 (400M reads) Paired End 50bp (PE50) + Bioinformatics Processing | 1980 | |
| NovaSeq X (1500M reads) Paired End 50bp (PE50) + Bioinformatics Processing | 3484 |
50M reads of paired end 50 (PE50) sequencing is recommended per sample. Multiplexing optimization depends on the factor used, for well characterized antibodies or sharp histone marks (for example: H3K4me3), 25 million reads may be sufficient per sample.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
ATAC-seq (assay for transposase activity)
Assay description
Assessment of the functional state of chromatin, can be achieved through the digestion of chromatin with Tn5 transposase followed by library preparation and sequencing. The Epigenomics core uses the OMNI-ATAC protocol as detailed in Corces et al. (Nature Methods, 2017)
Sample Requirement:
After submitting a service request in Agilent Crosslab (formerly iLab), please follow the protocol detailed below, label the Eppendorf tube with the Agilent CrossLab ID and the sample number (as in the serviece request) and bring samples after tagmentation to room A-427
Cell preparation
Make sure the cells are viable! This technique requires viability above 90% and preferably around 95%.
- Wash 50,000 cells with 1000 μl of ice-cold PBS. Centrifuge at 500 x g for 5 min at 4°C.
- Carefully remove most of the supernatant, leaving a little volume behind to avoid disturbing the pellet. Resuspend the pellet in 25 μl of ice cold 1X ATAC Buffer. Incubate 5 min on ice.
- Add 25 μl of ice cold ATAC-Detergent-buffer. Pipette up and down 3 times.
- Incubate the samples on ice for 3 min.
- Centrifuge at 500 x g for 10 min at 4°C.
- Remove the supernatant. Proceed immediately with the transposase reaction.
Tagmentation
- Resuspend the pellet in the following transposase mixture:
25 μl 2X TD Buffer (Illumina 20034197 or 20034198)
2.5 μl TDE1 (Illumina 20034197 or 20034198)
16.5 ul PBS
0.5 ul 1% Digitonin
0.5 ul 10% Tween-20
5 ul H2O - Incubate the reaction at 37°C for 30 min in thermomixer set to 500 rpm.
- Add 250 uls of Zymo DNA binding buffer to samples (5 fold).
- Store at -20oC until all the experimental samples are ready, then bring to the core. The DNA is stable for at least 2 weeks.
If your lab has Zymo columns and you want to proceed with clean up, please to do.
Remember to add as comment in the Crosslab/iLab submission that the tagmented DNA is in EB
- Load the mixture into a Zymo-Spin Column in a Collection tube.
- Centrifuge at full speed (>10,000 g) for 30 secs discard the flow through.
- Add 200 ul of DNA Wash buffer to the column and centrifuge for 30 seconds. Repeat the wash step. Do one extra 30 secs spin.
- Place the Zymo Spin Column into a new 1.5 ml tube. Elute with 22 μl of EB, and bring 20 μl to the core.
Reagents
2X ATAC-Buffer
| Reagent | Final Concentration | Volume for 5 ml |
| 1M Tris-HCl (pH 7.4) | 20 mM | 100 ul |
| 5M NaCl | 20 mM | 20 ul |
| 1M MgCl2 | 6 mM | 30 ul |
| Sterile water | NA | 4.85 ml |
- Digitonin - (Promega cat# G9441) Digitonin is supplied at 2% in DMSO.
- Dilute 1:1 with water to make a 1% stock solution. Avoid more than 5 freeze thaw cycles. Can be kept at -20°C for up to 6 months (100x)
- Tween-20 – (Sigma/Aldrich cas# 9005-64-5; cat# P9416-molecular biology grade). Supplied at 100%. Dilute to 10% and use at this concentration (100x stock). Store at 4°C.
- Igepal CA-630 – (Sigma/Aldrich cas# 9002-93-1; cat # I8896-molecular biology grade) Supplied at 100%. Dilute to 10% and use at this concentration (100x stock).
- Illumina - Tagment DNA TDE1 Enzyme and Buffer Kit (Small #20034197, Large #20034198)
- Zymo - DNA binding buffer (Zymo Research D4003)
On the day of the experiment use 2X ATAC-Buffer to prepare:
ATAC Buffer
| Reagent | Final Concentration | Volume for 0.05 ml (1 reaction) |
| 2X ATAC-Buffer | 1X | 25 ul |
| Sterile water | NA | 25 ul |
ATAC-Detergent Buffer
| Reagent | Final Concentration | Volume for 0.05 ml (1 reaction) |
| 2X ATACseq buffer | 1X | 25 ul |
| Igepal CA-630 10% | 0.20% | 1 ul |
| Tween 20 10% | 0.20% | 1 ul |
| Digitonin 1% | 0.02% | 1 ul |
| Sterile water | NA | 22 ul |
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| ATAC-seq (Omni) Library Prep (Tagmented Samples) | 100 | 140 |
| NextSeq 2000 (400M reads) Paired End 50bp (PE50) + Bioinformatics Processing | 1980 | |
| NovaSeq X (1500M reads) Paired End 50bp (PE50) + Bioinformatics Processing | 3484 |
50M reads of paired end 50 (PE50) sequencing is recommended per sample.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
Exome Capture
Assay description
The Agilent SureSelect platform is a solution-based system using 120-mer (biotinylated cRNA baits) to capture regions of interest, enriching them out of a NGS sonicated genomic library. Targeted regions can be interrogated for the purpose of identifying structural rearrangements and SNPs. [SureSelect PDF]
Sample Requirement:
Please submit a minimum of 12 samples. Submit 3ug of purified DNA, Nanodrop A260/280 ratio> 1.7; A260/230 =2.0-2.2
Epigenomics Core Quality Control:
dsDNA using Qubit Fluorometer, 0.8 % agarose gel to determine quality
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Exome Capture TruSeq Compatible Library Prep | 528 | 600 |
| HiSeq PE75 Paired End Clustering and 3 x 50 Sequencing Cycles | 2050 | 3000 |
1 lane of PE75 sequencing is recommended for commercially available capture libraries. To calculate the sequencing depth required for you species of interest, please use the Illumina coverage calculator.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
DNASeq (whole genome sequencing)
To continue availing of this service, please contact Dr. Jenny Xiang at the Genomics Resources Core Facility (GRCF).
[GRCF Service Request Portal]
An entire genome can be sequenced allowing SNP discovery, identification of copy number variations and chromosomal rearrangements [Nanokit]
Sample Requirement:
Submit 250 ng of genomic DNA, of molecular weight >40kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; RNA-free and at a concentration of ~20ng/ul
Epigenomics Core Quality Control:
dsDNA using Qubit Fluorometer, 0.8 % agarose gel to determine quality
[Click here for a detailed description of the QC]
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| TruSeq DNA-seq Library Prep | 200 | 300 |
Two lanes of PE75 or PE100 sequencing recommended for human DNAseq. To calculate depth required for your custom application, please use the Illumina coverage calculator.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
Targeted Resequencing (interrogation of genes specific for heme malignancies)
Assay description
Using Raindance’s single-molecule pico droplet PCR reaction three panels that contain genes specific for heme malignancies have been designed:
- Myeloid leukemia
- Chronic Lymphocytic Leukemia
- Lymphoma
This is a high-throughput technique, a minimum of 40 samples are required.
Sample Requirement:
Submit 500 ng of genomic DNA, of molecular weight >40kb, Nanodrop A260/280 ratio >1.7; A260/230=2.0-2.2; at ~20ng/ul
Epigenomics Core Quality Control:
Determination of concentration of double stranded DNA (dsDNA) using Qubit Fluorometer, Perkin Elmer Labchip GX or agarose gel to determine molecular weight.
| Service | Internal (WCM, WCM Qatar & Cornell U) Price |
External Academic Price |
|---|---|---|
| Raindance Library Prep | 200 | 400 |
| MiSeq 600 Sequencing Cycles v3 | 1500 | 2400 |
Please contact us if you wish to use this service.
Data ProcessingSequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are aligned to genomes available via Illumina's iGenome using the BWA-MEM aligner. This pipeline results in the following file types:
*.maxL.bam - The top/best non-filtered alignment for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.merged.bam - all alignments (including best and multiple alignments) for each read in the widely accepted BAM format (a binary version of the SAM format) for each sample.
*.maxL.bam.bai, *.merged.bam.bai - Index files (.bai files) for each sample which allow for easier viewing of the bam files in genome browsers such as UCSC Genome Browser) or IGV).
*-metrics.log - Summary metrics such as adapter trimming and alignment rates.
Single Cell Gene Expression
Assay description
We are currently offering 10x Genomics applications for our single cell gene expression assays using new Next GEM chips [PDF]
Single cell assays require prior consultation and coordination with the core. Please contact us at epicore-sc-coord@med.cornell.edu at least three weeks before dropping off your samples to schedule a time. We currently only accept external customers coming from within a mile radius of our facility.
The 10x Genomics Chromium Single Cell Expression Solution provides high-throughput, single cell expression measurements that enable discovery of gene expression dynamics and molecular profiling of individual cell types. This is also available in conjunction with Feature Barcoding (Cell Surface Protein, CRISPR Screening or Custom). The protocol requires a suspension of viable single cells as input. Minimizing the presence of cellular aggregates, dead cells, non-cellular nucleic acids and potential inhibitors of reverse transcription is critical to obtaining high quality data.
Please consult with us if you are interested in 10X Genomics Cell Multiplexing
Sample Requirement:
- The total number of cells required in the suspension used as input is determined by the desired cell recovery target (between 500-10000 cells); required sequencing read depth depends on the desired cell target. Please check user guide (page 10 and page 41 respectively) for details. [Library Prep User Guides]
- Given the variety of cells and sample types, general guidelines for sample preparation need to be optimized by each customer. Please check the 10X Genomics Single Cell Gene Expression Sample Prep Guide for information on how to prepare cells.
- We prefer to process up to 4 samples at a time due to workflow constraints and currently only use V3.1 chemistry.
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Chromium Single Cell Library Prep (1 sample) | $2230 | $2849 |
| Chromium Single Cell Library Prep (2 samples) | $4224 | $5084 |
| Chromium Single Cell Library Prep (3 samples) | $5880 | $6933 |
| Chromium Single Cell Library Prep (4 samples) | $7540 | $9016 |
| Feature Barcoding Library Prep (per sample) | $250 | $390 |
| NovaSeq (1.25B reads) + Bioinformatics Processing | $2000 | $3100 |
We sequence Chromium single cell samples on a pair end flow cell (28-10-10-90) with a 2x50 cycles kit. Chromium recommends a minimum of 20,000 reads per cell for gene expression (Example: Samples containing from 2000-10,000 cells would require from 40-200 million reads per sample). Please contact us at epigenomicscore@med.cornell.edu to discuss the appropriate number of reads required for your experiments.
Data Processing
The sequencing data will be demultiplexed and post-processed using custom pipelines provided by 10x Genomics.
Feature Barcoding (Cell Surface Protein, CRISPR Screening or Custom) and Cell Multiplexing assays require additional information for post-processing with cellranger (for collating the feature counts with the gene expression counts).
Please download and fill out our form for Feature Barcoding data: FeatureBarcodingInput.xlsx (Replace the examples with representation for your samples)
Below please find a description of the columns:
| Sample Name | The name you have used for the samples while submitting them in iLab |
| Feature Name | Human-readable name for this feature. Must not contain whitespace. This name will be displayed in Loupe Cell Browser. |
| Read (R1 or R2) | Specifies which RNA sequencing read contains the Feature Barocde sequence. Must be R1 or R2. Note: in most cases R2 is the correct read. |
| Pattern / Position of Barcode | The pattern column can be made up of a combination of these elements: 5P: denotes the beginning of the read sequence. May appear 0 or 1 times, and must be at the beginning of the pattern. Only 5P or 3P may appear, not both. 3P: denotes the end of the read sequence. May appear 0 or 1 times, and must be at the end of the pattern. N: denotes an arbitrary base. A, C, G, T: denotes a fixed base that must match the read sequence exactly. (BC): denotes the Feature Barcode sequence as specified in the sequence column of the feature reference. Must appear exactly once in the pattern. |
| Sequence | Nucleotide barcode sequence associated with this feature. E.g., antibody barcode or sgRNA protospacer sequence. |
The 10X Genomics Feature Barcoding Web Page has detailed information about the post processing of feature barcoding data and required input for cellranger.
Please download and fill out the following form for Cell Multiplexing data: CellplexInput.xlsx (Replace the examples with representation for your samples)
Single Cell Immune Profiling
Assay description
We are currently offering 10x Genomics applications for our single cell immune profiling assays using new Next GEM chips [PDF]
Single cell assays require prior consultation and coordination with the core. Please contact us at epicore-sc-coord@med.cornell.edu at least three weeks before dropping off your samples to schedule a time. We currently only accept external customers coming from within a mile radius of our facility.
The Chromium Single Cell Immune Profiling Solution
The Chromium Single Cell Immune Profiling Solution is a comprehensive approach to simultaneously examine the cellular context of the adaptive immune response and immune repertoires of hundreds to tens of thousands of T and B cells in human or mouse on a cell-by-cell basis. With the addition of Feature Barcoding Technology, you can now also detect and analyze additional cellular readouts such as cell surface markers and antigen specificity to enhance immune cell phenotyping and study dynamic interactions between lymphocytes and target cells.
Sample Requirement:
The protocol requires a suspension of viable single cells as input. Minimizing the presence of cellular aggregates, dead cells, non-cellular nucleic acids and potential inhibitors of reverse transcription is critical to obtaining high quality data.
- The total number of cells required in the suspension used as input is determined by the desired cell recovery target (between 500-10000 cells); required sequencing read depth depends on the desired cell target. Please check user guide (page 10 and page 41 respectively) for details. [Library Prep User Guide]
- Given the variety of cells and sample types, general guidelines for sample preparation need to be optimized by each customer. Please check the 10X Genomics Single Cell Immune Profiling Sample Prep Guide for information on how to prepare cells.
- We can process up to 4 samples at a time.
The library preparation prices below are for the 5 prime gene expression assay as well as either a T or a B cell V(D)J assay.
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| Chromium Single Cell Library Prep (1 sample) | $2342 | $2692 |
| Chromium Single Cell Library Prep (2 samples) | $4300 | $4730 |
| Chromium Single Cell Library Prep (3 samples) | $6000 | $6600 |
| Chromium Single Cell Library Prep (4 samples) | $7600 | $8400 |
| NovaSeq (1.25B reads) + Bioinformatics Processing | $2000 | $3100 |
We sequence Chromium single cell samples on a pair end flow cell (28-10-10-91) with a 2x50 cycles kit. Chromium recommends 50,000 reads per cell with a minimum of 20,000 reads per cell for gene expression (Example: Samples containing from 2000-10,000 cells would require from 100-500 million reads per sample). The T or B cell specific V(D)J assay requires 5000 reads per cell. Please contact us at epigenomicscore@med.cornell.edu to discuss the appropriate number of reads required for your experiments.
Data Processing
The sequencing data will be demultiplexed and post-processed using custom pipelines provided by 10x Genomics.
Single Cell Epigenomics
Assay description
We are currently offering Single Cell ATAC (NextGEM) as well as Single Cell RRBS (96 well plate protocol with or without SMART-Seq single cell gene expression).
Single cell assays require prior consultation and coordination with the core. Please contact us at epicore-sc-coord@med.cornell.edu at least three weeks before dropping off your samples to schedule a time.
The Chromium Single Cell ATAC (Assay for Transposase Accessible Chromatin) Solution accelerates the understanding of the regulatory landscape of the genome, thereby providing insights into cell variability. The high-throughput chromatin profiling of single cells in parallel allows researchers to see how chromatin compaction and DNA-binding proteins regulate gene expression at high resolution.
Sample Requirement:
- The total number of nuclei required in the suspension used as input is determined by the desired cell recovery target (between 500-10,000 cells); required sequencing read depth depends on the desired cell target. Chromium recommends 25,000 reads per nuclei (Example: 4 samples at 10,000 cells can be sequenced on one NovaSeq6000 SP flowcell, sequencing recipe 50-8-16-50) [Library Prep User Guide]
- Given the variety of cells and sample types, general guidelines for sample preparation need to be optimized by each customer. Please check the 10X Genomics Single Cell ATAC Sample Prep Guide for information on how to prepare cells.
- We can process up to 4 samples at a time and can only accept samples from local/internal clients.
| Service | Internal Price |
|---|---|
| Chromium Single Cell ATAC Library Prep (1 sample) | $3000 |
| Chromium Single Cell ATAC Library Prep (2 samples) | $4000 | Chromium Single Cell ATAC Library Prep (6 samples) | $12000 |
Single Cell RRBS with SMART-Seq Gene Expression
We developed an in-house Single Cell RRBS protocol as a 96 well plate assay which can be performed with or without SMART-Seq gene exression in parallel. Please contact us at epigenomicscore@med.cornell.edu for further information and to discuss the details of your project.
Data Processing
The sequencing data from Chromium Single Cell ATAC will be demultiplexed and post-processed using custom pipelines provided by 10x Genomics.
The sequencing data from our Single Cell RRBS assays will be demultiplexed and post-processed in the same manner as our RRBS samples. Please click on the ERRBS section in the right hand menu for details.
Chromium Long Range Sequencing
Assay description
We are currently offering 10x Genomics applications for our long range genomic assay.
The Chromium Long Range Genome Solution provides long range information on a genome-wide scale, including variant calling, phasing and extensive characterization of genomic structure. Applications include interrogating heterogeneous cell populations, resolving phasing information and detecting structural variations. Please contact us at epigenomicscore@med.cornell.edu for further information and to discuss the details of your project.
Data Processing
The sequencing data will be demultiplexed and post-processed using custom pipelines provided by 10x Genomics.
Spatial Transcriptomic Profiling
Assay Description
We are currently offering Visium CytAssist Spatial Gene Expression for FFPE, H&E workflow for mouse and human tissue samples. This assay allows mapping of the transcriptome within morphological context.
Spatial Gene Expression assays require prior consultation and coordination with the core. Please contact us at epicore-sc-coord@med.cornell.edu at least three weeks before dropping off your samples to schedule a time.
General Workflow
Phase one, performed by the researcher or through a Histology Core:
- FFPE RNA quality control (recommended DV200 > 30%)
- Preliminary H&E staining to determine the region of interest
- Microtome sectioning and tissue placement on a standard histology slide (for details refer to the Sample Requirements section below and the Planning a CytAssist Visium FFPE Spatial Gene Expression experiment [PDF] guide)
- Deparaffinization, H&E staining and Imaging
- Probe hybridization, ligation, extension and release and library construction
- Library QC
- Sequencing and Data Processing
Sample Requirements
- Samples are 3-10µm-thick FFPE tissue sections on standard histology slides (“Tissue Slides”) (minimum slide size 24.8 x 74.4mm / maximum 25.3 x 76.2mm)
- The region of interest (6.5mm2 or 11mm2) must be within an allowable area on the “Tissue Slide” (for size and location of the allowable area refer to Planning a CytAssist Visium FFPE Spatial Gene Expression experiment PDF file)
- Freshly prepared slides have to dry at 42C for 3h and stored at RT in a desiccator for up to two weeks.
- The protocol has been tested and developed for archived FFPE slides as well.
Sample Submission
- Login to your Agilent CrossLab/iLab account and select the Epigenomics core facility
- Initiate a request for Spatial Transcriptomics. For details on how to submit a Spatial Transcriptomics request refer to the last paragraph of the Planning a CytAssist Visium FFPE Spatial Gene Expression experiment [PDF] guide.
- Each “Tissue Slide” must be labeled in accordance with the iLab submission form and the annotated preliminary H&E images (see here below).
- On the iLab request page, upload a PDF file with the annotated low-resolution H&E image of the entire tissue section for each of the submitted “Tissue Slide”. Each H&E image will have to clearly show the chosen 6.5mm2 or 11mm2 region of interest and must be labeled with the corresponding “Tissue Slide” identifier.
- Tissue Slides can be transferred to the Epigenomic Core in a sealed 50ml falcon tube or slide box with few Drierite granules or small silica gel packets at RT. The Epigenomics Core has a desiccator chamber for the two-weeks RT storage, if necessary.
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| CytAssist 6.5mm2 (library prep and sequencing for two Capture Areas on a Visium slide) | $4700 | $5500 |
| CytAssist 11mm2 (library prep and sequencing for two Capture Areas on a Visium slide) | $11220 | $12903 |
Submit a minimum of 2 tissue slides, or multiples of 2. As many tissues as can fit may be added to the allowable area on a tissue slide. Libraries are sequenced on a pair end flow cell (28-10-10-50) with a 2x50 cycles kit. 10x Genomics recommends a minimum of 25,000 reads per spot covered by tissue. For a CytAssist 6.5mm2, 125M reads per square are needed. For CytAssist 11mm2, 400M reads per square are needed.
Data Processing
The sequencing data from Visium Spatial Gene Expression will be demultiplexed and post-processed along with microscope generated images using the spaceranger software provided by 10x Genomics.
Single Cell Spatial Profiling
Assay Description
We are currently offering multiomic single-cell spatial profiling via the CosMx Spatial Molecule Imager from NanoString for FFPE and Fresh Frozen samples/slides. This assay allows high plex profiling of transcriptomics and proteomics within morphological context at sub-cellular resolution.
Single cell spatial profiling assays require prior consultation and coordination with the core. Please contact us at epicore-sc-coord@med.cornell.edu at least three weeks before dropping off your samples to schedule a time.
Sample Requirements
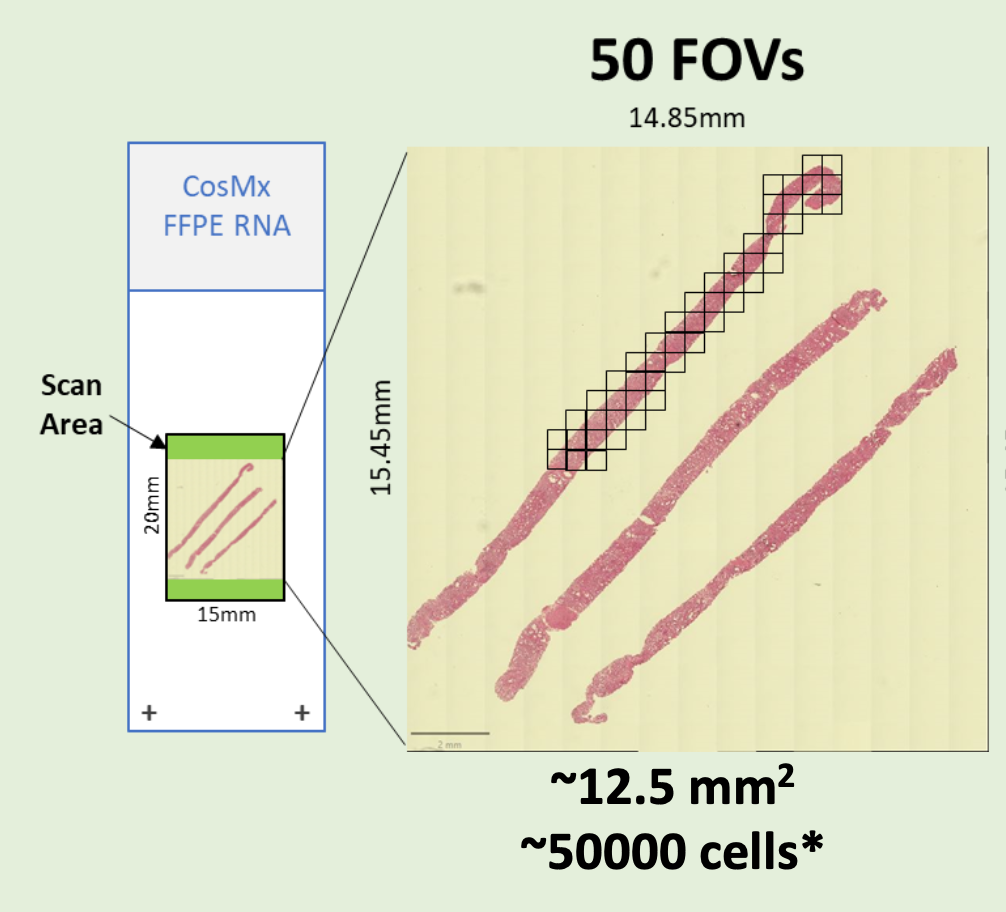
- FFPE microtome (RNA and Protein) or fresh frozen (RNA) sections on Nanostring-validated positively charged histology slides are required. Please review this detailed PDF to plan your CosMx experiment
- Preliminary sectioning, and H&E staining on parallel sections are required to determine the regions of interest and assign the prospective “Field of View” (FOVs) to be scored. Each “Field of View” (FOV) corresponds to a single CosMx 23x/1.1 NA objective acquisition area. FOVs are square-shaped, with the sides parallel to the slide edges. (It is not possible to rotate the FOVs to better fit the regions of interest.)
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
|---|---|
| Up to 150 FOVs per slide and 2-slides/run - RNA 1000-plex | $5146 |
| Up to 150 FOVs per slide and 2-slides/run - Protein 64-plex | $3680 |
| Up to 75 FOVs per slide and 4-slides/run - RNA 1000-plex | $3831 |
| Up to 75 FOVs per slide and 4-slides/run - Protein 64-plex | $2568 |
Submit a minimum of 2 histology slides, or multiples of 2.
Data Processing
The imaging data from CosMx will be processed in NanoString's AtoMx SIP and shared there-in. After running the foundational pipelines in AtoMx, we will also create and share a Seurat object for downstream analysis.
RNASeq (transcriptome analysis)
To continue availing of this service, please contact Dr. Jenny Xiang at the Genomics Resources Core Facility (GRCF).
[GRCF Service Request Portal]
To understand the dynamic state of the cell transcriptome, the core provides a range of RNA sequencing library preparations that can assess total RNA, mRNA and noncoding RNA in higher eukaryotes:
-
TruSeq Stranded mRNA:
polyA selection followed by library preparation is the standard and most widely used method for quantifying mRNA expression. Our protocol retains the strand orientation of the transcripts. [MORE]
Sample Requirement: Submit a minimum of 250 ng of purified RNA, at a concentration of 50 ng/ul; A260/280 > 2, picogreen quantification or bioanalyzer trace, RNA integrity number (RIN) >8.0
Epigenomics Core Quality Control: RNA Nano Bioanalyzer chip to determine RIN number and picogreen quantification.
-
Ultra Low Input RNA:
mRNA profiling of very low amounts of input material (<20ng) that is based on polyA selection using the Clontech SMArter technology. Often used for clinical samples or expression profiling of few cells (e.g. neurons, stem cells).
Sample Requirement: Submit a minimum of 20 ng of purified RNA, at a concentration of 5 ng/ul; A260/280 > 2, picogreen quantification or bioanalyzer trace, RNA integrity number (RIN) >8.0 OR 1000 cells in 10ul of Clontech 1X Single Cell Lysis buffer (cat # 635013)
Epigenomics Core Quality Control: RNA Pico Bioanalyzer chip to determine RIN number and picogreen quantification.
-
TruSeq Stranded Total RNA:
rRNA depletion followed by strand-specific library preparation. Generally more applicable towards simultaneous profiling of non-coding transcripts and mRNA although requires more input material. [MORE]
Sample Requirement: Submit a minimum of 250 ng of purified RNA, at a concentration of 50 ng/ul; A260/280 > 2, picogreen quantification or bioanalyzer trace, RNAs with an RNA integrity number (RIN) < 8 can be used
Epigenomics Core Quality Control: RNA Nano Bioanalyzer chip and picogreen quantification.
-
TruSeq RNA [DISCONTINUED]:
Please use TruSeq-stranded mRNA instead.
Agilent's Guide to Interpreting Bioanalyzer Results
Some studies that have used our RNASeq service: Kuo PY. et al. 2014, Satyaki PR. et al. 2014, Pimentel H. et al. 2014
| Service | Internal Price (WCM, WCM Qatar & Cornell U) |
External Academic Price |
|---|---|---|
| TruSeq RNA-seq (polyA) Library Prep | 185 | 300 |
| TruSeq Stranded RNA-seq (polyA) Library Prep | 170 | 225 |
| Ultra Low Input RNA-seq (polyA) Library Prep | 280 | 370 |
| TruSeq Total RNA-seq (stranded) Library Prep | 300 | 400 |
| HiSeq SR50 Single Read Clustering and 1 x 50 Sequencing Cycles | 1100 | 1600 |
| HiSeq PE50 Paired End Clustering and 2 x 50 Sequencing Cycles | 1650 | 2200 |
Sequencing depth required depends on your application.
For human transcriptome differential gene expression we recommend 4 samples per lane of SR50 sequencing.
For rare splicing events, translocations and some other experiments PE50 sequencing is recommended.
To calculate depth required for your custom application, please use the Illumina coverage calculator.
Sequence data (base call files or bcl files) generated from the sequencer are demultiplexed and converted to FASTQ files using the Illumina bcl2fastq software.
Your raw data will be available for download as a tar compressed archive (Sample_*.tar) of gzipped FASTQ files for each sample. Raw data can be post-processesed upon request.
FASTQ files generated as described above are adapter trimmed and aligned to genomes available via Illumina's iGenome using the STAR aligner. Only raw reads that pass Illumina's purity filter are aligned. This pipeline results in the following file types:
*.bam - Upon alignment (if requested) the aligned data processed by STAR aligner is in the widely accepted BAM format (a binary version of the SAM format).
*.bai - This is an index file for your BAM alignments and allows certain browsers (such as the IGV browser) to better view the .bam file.
*-SJ.out.tab - The high confidence collapsed splice junctions in tab-delimited format. Only junctions supported by uniquely mapping reads are reported.
*-Log.final.out - A text file containing the STAR aligner generated summary statistics for the alignment of each sample.
Bioinformatics Support
The Epigenomics Core Facility provides data analysis services and consultation on a per project basis. These include but are not limited to:
- RNASeq differential expression
- ERRBS differential methylation
- ChIPSeq peak calling and differential binding
- Functional annotation and analysis
- Single Cell and Spatial Profiling
- Custom alignments
Please note that some bioinformatics support is included in the price for some of our services. For example:
- Quality control statistics for your samples
- 2 years data storage on our backed up servers with unlimited password protected access for sequencing projects
- Illumina iGenome alignments (if available and when requested for internal customers)
- Methylation calling for DNA methylation sequencing projects (where genomes are available)
- Methylation ratios for Epityper mass array projects
- Splice junction tables for RNASeq alignments
Please review the Data Analysis and Retrieval section of our Frequently Asked Questions page for further details.
| Service Type | Internal Price (WCM, WCM Qatar & Cornell U) | External Academic Price |
|---|---|---|
| Data analysis support / hour | $100 | $150 |
Related Publications:
- Zhang, T. et al. Blood Cancer J. 2017. Sep 8; 7 (9), e606 [PubMed]
- Bayliss, J., Mukherjee P. et al. Sci Transl Mel 2016. Nov 23; 8 (366), 366ra161 [PubMed]
- Garrett-Bakelman, FE. Sheridan, C. et al. J Vis Exp. 2015. Feb 24; (96) [PubMed]
- Marcinkiewicz, KM. et al. J Cell Physiol. 2014. 229(10):1405-16. [PubMed]
- Rapaport, F. et al. Genome Biology. 2013. Sep; 14(9):R95. [PubMed]
- Janovitz, T. et al. Journal of Virology. 2013. Aug; 87(15):8559-68. [PubMed]
- Oh, JE. et al. Translational Psychiatry. 2013. Jan 22;3:e218. [PubMed]

